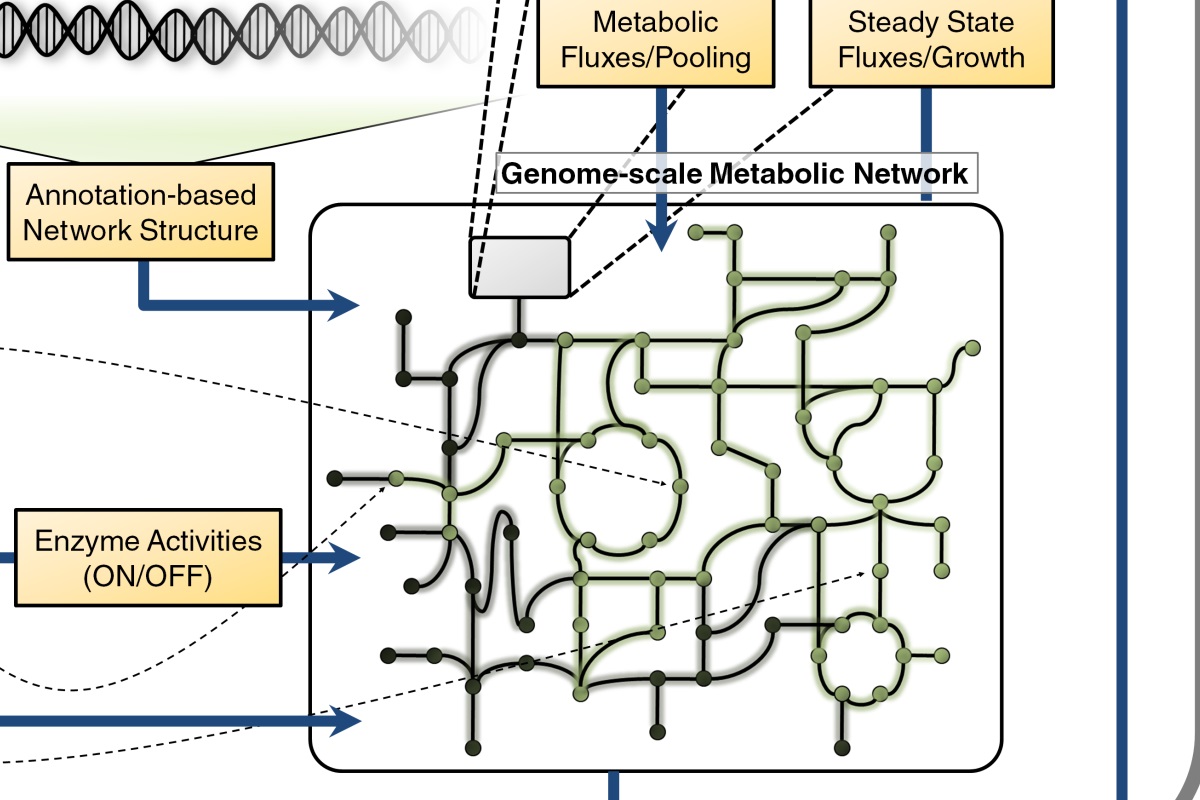
Methanosarcina acetivorans reconstruction
In collaboration with Bill Metcalf’s lab at the University of Illinois at Urbana-Champaign, we are sequencing 20 novel Methanosarcina strains. The genomes are being analyzed to learn more about the evolution of metabolic and regulatory networks to identify environmental patterns in their evolution. As part of this analysis, we have constructed a genome scale metabolic model of Methanosarcina acetivorans C2A, a model archaeon. Along with our updated model of M. barkeri, this model serves as a platform to enhance our understanding of methanogen metabolism, a vital part of the global carbon cycle.
Data and Software File(s):
iMB745 SBML model


 hood-price.isbscience.org/research/methanosarcina-acetivorans-reconstruction/
hood-price.isbscience.org/research/methanosarcina-acetivorans-reconstruction/