Kinetic modeling of Parkinson’s Disease
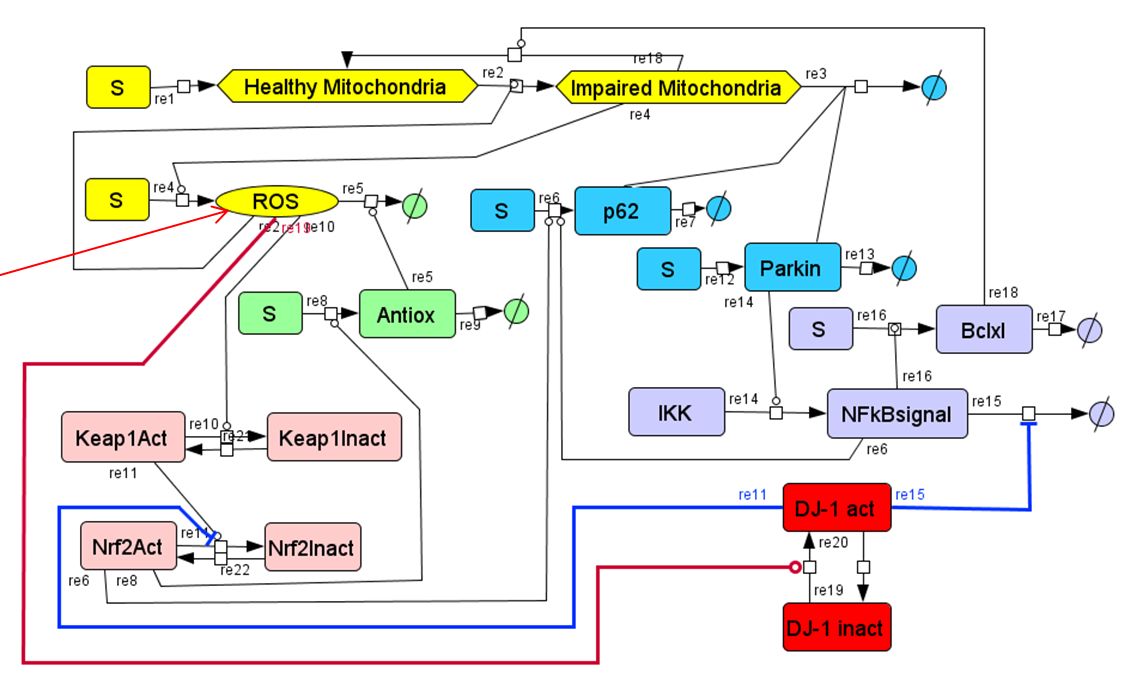
In Parkinson’s disease (PD), the activities of several components of the reactive oxygen species (ROS)-activated regulatory network are altered. This makes the cell susceptible to ROS damage. In the case of dopaminergic neurons, this effect can be particularly severe. In order to understand the functioning of the ROS-activated regulatory network in normal function and disease, we have built a kinetic model using bottom-up mechanism-based ODE kinetic modeling. For this approach, we start with the known network of interactions between different species (proteins, mRNA, DNA and other biomolecules). The change of each species’ concentrations is expressed through balance equations and rate equations. Then, the change of each species’ concentration is integrated into an ODE (Ordinary Differential Equations) system. Solving this ODE system allows simulating the kinetic behavior of modeled biochemical network. Our model includes 39 species and 45 reactions, with 56 kinetic parameters, either fitted to experimental data or obtained from the literature. Our model allows the simulation of PD-related ROS generation and mitochondrial damage. We are currently improving the kinetic model so that it can be used to better understand the pathophysiology of PD and to guide the development of novel mitochondria-related PD treatments.


 hood-price.isbscience.org/research/dynamic-network-modeling/
hood-price.isbscience.org/research/dynamic-network-modeling/